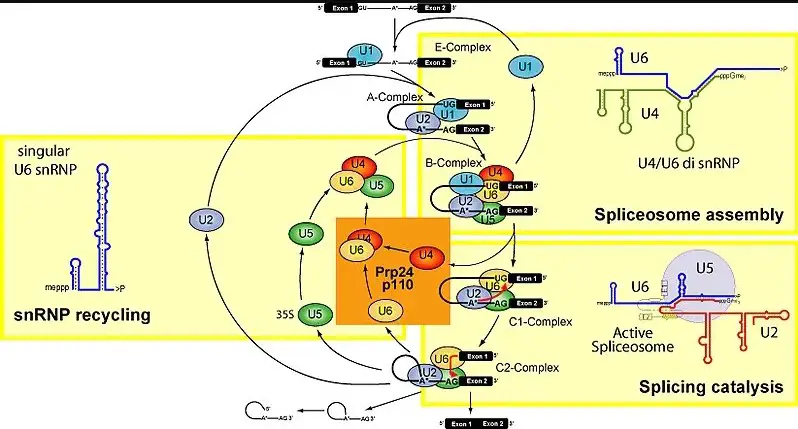
Gene expression is a sophisticated cellular process, fundamental to all living organisms, involving multiple molecular players. Among these, small nuclear RNAs (snRNA) and small nuclear ribonucleoproteins (snRNP) are pivotal in the RNA splicing phase, ensuring the accurate processing of precursor messenger RNA (pre-mRNA) into mature RNA. These components are not only crucial for the proper expression of genes but also play significant roles in the regulation of genetic activity across various biological systems.
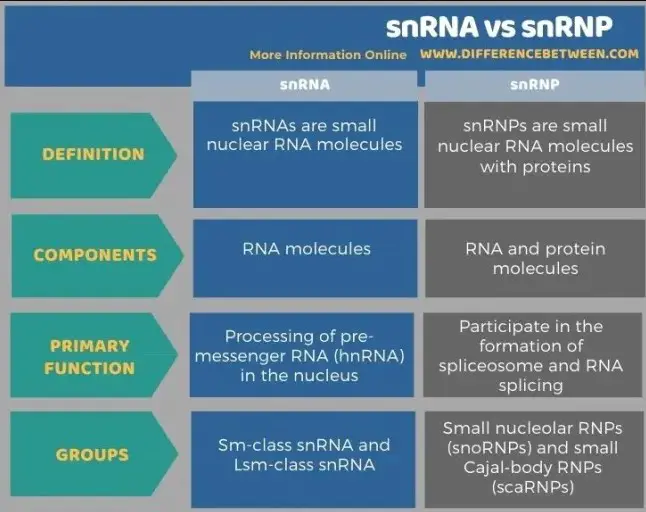
snRNAs are small RNA molecules that form complexes with proteins to create snRNPs, which in turn are essential components of the spliceosome—the cellular machinery responsible for editing pre-mRNA. The primary difference between snRNA and snRNP is that snRNA refers to the RNA component alone, while snRNP includes both the snRNA and its associated proteins, making it a functional unit within the spliceosome.
Understanding the distinctions between snRNA and snRNP is not merely academic; it has profound implications in biomedical research and therapeutic development. The precise functioning of these molecules is critical for cell health and can influence a variety of genetic disorders. By exploring their structural and functional differences, researchers continue to uncover the intricate details of gene expression and its impact on health and disease.
SnRNA Basics
Definition and Role
Small nuclear RNA (snRNA) is a crucial component of the molecular machinery within eukaryotic cells. These RNA molecules, typically ranging from 100 to 300 nucleotides in length, are not involved in coding for proteins but play a pivotal role in the post-transcriptional modification of pre-messenger RNA (pre-mRNA), a process essential for the accurate formation of messenger RNA (mRNA).
What is SnRNA?
snRNA molecules reside within the nucleus of the cell. They are best known for their function in the spliceosome, the complex responsible for RNA splicing—the editing of pre-mRNA by removing introns and connecting exons. This editing is vital for the generation of viable mRNA that will later translate into proteins.
Functions in the Cell
The primary function of snRNA is to ensure the fidelity of gene expression. This is accomplished by:
- Identifying splice sites: snRNA sequences recognize specific nucleotide sequences on the pre-mRNA that designate where splicing should occur.
- Catalyzing chemical reactions: Some snRNAs, like U6, are catalytic and actively participate in the splicing reactions.
Types and Features
Common Types of SnRNA
There are several types of snRNAs, with U1, U2, U4, U5, and U6 being among the most studied due to their direct involvement in the spliceosome. Each type has a unique sequence and structure, enabling it to bind specifically to certain sequences or structures within the pre-mRNA.
Structural Characteristics
snRNAs are characterized by their small size and distinct secondary structures, which include stem-loops and single-stranded regions. These structures are critical for binding with proteins to form snRNPs and for the snRNA’s function in the spliceosome.
snRNP Basics
Definition and Components
Small nuclear ribonucleoproteins (snRNPs) are complexes formed by the combination of snRNA and protein molecules. These complexes are fundamental constituents of the spliceosome.
What is snRNP?
An snRNP consists of one or more snRNA molecules along with several proteins. This composition not only stabilizes snRNA but also enhances its functionality in RNA splicing. Each snRNP is named after its snRNA component—U1 snRNP contains U1 snRNA, and so on.
Core Components of snRNP
The core components of an snRNP include:
- snRNA: Provides the template for recognizing specific nucleotide sequences in the pre-mRNA.
- Sm proteins: A heptameric ring of proteins that binds specifically to the snRNA, forming a stable snRNP.
- Additional proteins: Depending on the type of snRNP, additional proteins may be involved to aid in splicing or regulation.
Function and Importance
Role in Spliceosome
snRNPs are the building blocks of the spliceosome, which is assembled on the pre-mRNA substrate in a stepwise manner. The assembly process involves multiple snRNPs that recognize and bind to specific sequences of the pre-mRNA, ensuring accurate splicing.
Contribution to RNA Splicing
The contribution of snRNPs to RNA splicing includes:
- Complex assembly: Each snRNP has a specific role in the assembly and rearrangement of the spliceosome complex.
- Splice site recognition and activation: snRNPs help identify the correct splice sites and activate them for splicing.
- Catalysis of splicing reactions: Certain snRNPs, such as U6, are directly involved in the catalysis of splicing reactions.

Key Differences
Structural Distinctions
Composition Comparison
snRNA and snRNP differ primarily in their composition. While snRNA consists solely of RNA, snRNP includes both snRNA and a set of associated proteins. These proteins, often referred to as Snurportin and Sm proteins, are crucial for the snRNA’s integration into the spliceosome and its overall stability within the nucleus.
Molecular Differences
On a molecular level, snRNAs are relatively simple structures composed of RNA nucleotides that form secondary structures like hairpins and loops. In contrast, snRNPs are more complex, featuring a core snRNA molecule surrounded by multiple proteins. This arrangement allows snRNPs to have enhanced functional capabilities, including improved recognition sites for splicing and increased stability under cellular conditions.
Functional Variances
Role in Gene Expression
snRNA’s role in gene expression is primarily through its involvement in the splicing process, where it helps ensure that splicing occurs at the correct locations. snRNP, however, contributes more dynamically by also regulating the assembly and disassembly of the spliceosome complex, thus playing a critical role in the temporal and spatial control of gene expression.
Impact on Splicing Mechanisms
The presence of protein components in snRNPs allows for a more nuanced interaction with the pre-mRNA substrate. This interaction is crucial for the catalytic steps of splicing, where precise cuts and ligation determine the mRNA outcome. The proteins in snRNPs can recognize and respond to the complex chemical environment of the spliceosome, facilitating accurate splicing under a variety of cellular conditions.
Biological Significance
In Genetic Regulation
Both snRNA and snRNP are pivotal in genetic regulation. By ensuring accurate splicing, they maintain the integrity of the genetic information translated into proteins, crucial for cellular function and development. Errors in their function can lead to mis-splicing, which is often associated with various genetic disorders and diseases.
SnRNA and snRNP in Gene Control
The precise control of gene expression by snRNA and snRNP is fundamental to cellular differentiation and organismal development. These molecules ensure that genes are turned on and off at the right times and that the mRNA transcripts are accurately edited before translation, critical for the correct expression of genetic information.
Medical Perspectives
Implications in Diseases
Misfunction of snRNAs and snRNPs can lead to a variety of diseases, including spinal muscular atrophy and retinitis pigmentosa. These conditions are often linked to mutations that affect snRNA or the proteins associated with snRNPs, disrupting normal splicing and leading to faulty gene expression.
Potential Therapeutic Targets
Given their central role in gene expression, both snRNA and snRNP are considered promising targets for therapeutic interventions. Developing molecules that can modulate the function of snRNAs or snRNPs might allow for the correction of splicing defects at the molecular level, offering new avenues for treatment of genetic diseases.
Research and Innovations
Recent Studies
Recent advancements in the study of snRNA and snRNP have shed light on their complex roles in cellular physiology and disease. Innovative techniques like CRISPR-Cas9 and single-molecule microscopy have allowed researchers to observe the real-time dynamics of snRNPs within the spliceosome, leading to a deeper understanding of their function and regulation.
Advances in SnRNA and snRNP Research
Studies have increasingly focused on how snRNAs and snRNPs interact with other components of the gene expression machinery. These interactions are crucial for the development of more effective gene therapies, particularly those aimed at correcting splicing errors at their root.
Future Prospects
Emerging Technologies and Their Potentials
The future of snRNA and snRNP research looks promising with the development of technologies that allow for precise editing of RNA and protein components. Potential technologies include RNA trans-splicing and protein engineering, which could revolutionize the treatment of diseases caused by splicing errors. The continued exploration of these molecules is likely to yield substantial benefits for genetic medicine, enhancing our ability to treat and possibly cure many genetic disorders.
Frequently Asked Questions
What is snRNA?
Small nuclear RNA (snRNA) is a type of non-coding RNA that plays a critical role in the splicing of precursor messenger RNA in the nucleus of eukaryotic cells. It helps ensure the accurate removal of introns and joining of exons, which is essential for the synthesis of correct and functional messenger RNA.
How does snRNP contribute to splicing?
snRNP, or small nuclear ribonucleoprotein, contributes to the splicing process by forming part of the spliceosome complex. It uses the snRNA as a guide to recognize specific sequences within the pre-mRNA, thereby facilitating the precise excision of introns and the ligation of exons.
What are the main differences between snRNA and snRNP?
The main difference between snRNA and snRNP is that snRNA is the RNA component itself, while snRNP refers to the complex of snRNA bound to specific proteins. This complex is crucial for the snRNA’s stability and functionality within the spliceosome.
Why are snRNA and snRNP important in genetic regulation?
snRNA and snRNP are vital for the regulation of gene expression as they ensure the accurate splicing of pre-mRNA. Faulty splicing can lead to genetic disorders and diseases, making these molecules critical for cellular function and health.
Conclusion
The exploration of snRNA and snRNP provides essential insights into the molecular mechanics of gene expression. These components not only serve as critical elements in RNA splicing but also offer potential targets for therapeutic interventions in genetic diseases. Their distinct yet interrelated functions underline the complexity and precision required in genetic regulation.
Further research into the differences and interactions between snRNA and snRNP could lead to breakthroughs in our understanding of cellular processes and the development of new medical treatments. As science continues to unravel the intricate dance of these molecules, the potential for innovative solutions to complex genetic disorders remains promising.