RNA polymerase plays a pivotal role in the transcription process, translating DNA sequences into messenger RNA, a crucial step in the flow of genetic information within a cell. This enzyme’s functionality is foundational to both prokaryotic and eukaryotic organisms, facilitating the complex process of gene expression. Despite sharing a common purpose, the RNA polymerases in these two cell types exhibit significant differences in structure, regulation, and mechanism of action.
The primary difference between prokaryotic and eukaryotic RNA polymerase lies in their complexity and regulation. Prokaryotic organisms have a single RNA polymerase that manages the transcription of all types of RNA, whereas eukaryotic cells contain three main types of RNA polymerases (I, II, and III), each responsible for transcribing different sets of genes. This distinction reflects the more complex control over gene expression required in eukaryotic cells, which have a compartmentalized cell structure and more intricate developmental processes.
Furthermore, eukaryotic RNA polymerases require a variety of accessory proteins or factors to initiate transcription, reflecting the higher level of regulatory control in these organisms. The presence of introns in eukaryotic genes and the associated splicing mechanism for mRNA processing further complicate the eukaryotic transcription process. The differences in RNA polymerase between prokaryotes and eukaryotes underscore the evolutionary adaptations to their respective cellular environments, highlighting the complexity of eukaryotic gene regulation and the efficiency of prokaryotic transcription.
Core Concepts
RNA Polymerase Defined
Definition and Role
RNA polymerase is an enzyme that plays a pivotal role in the process of transcription, where it reads the DNA sequence of a gene and synthesizes a complementary RNA strand. This process is fundamental for the expression of genes, allowing the information stored in DNA to be translated into proteins that perform essential functions in living organisms.
Prokaryotic vs Eukaryotic Cells
Key Differences
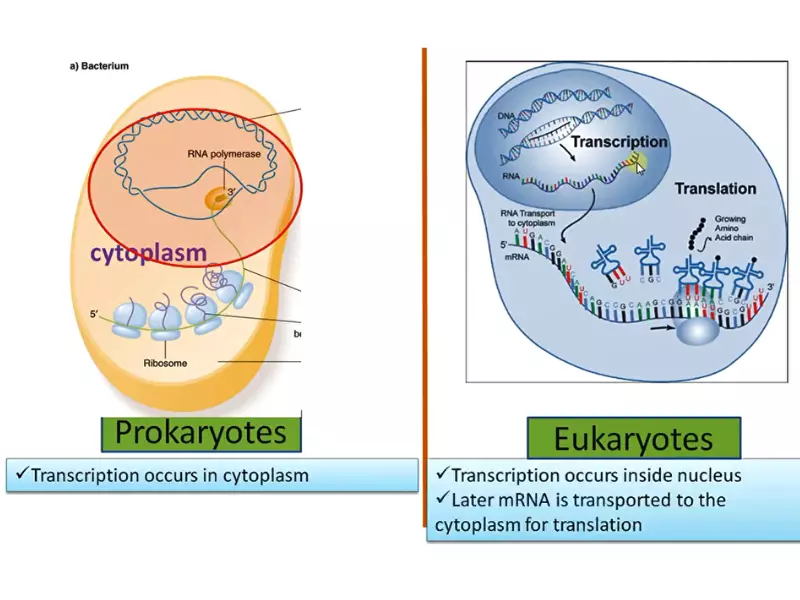
The primary distinction between prokaryotic and eukaryotic cells lies in their cellular organization. Prokaryotic cells, which include bacteria and archaea, are simpler and lack a nucleus. Eukaryotic cells, found in plants, animals, fungi, and protozoa, are more complex, featuring a nucleus and various other organelles. This fundamental difference extends to the RNA polymerase enzyme, where the structure, function, and regulation differ significantly between prokaryotes and eukaryotes.
RNA Polymerase in Action
Prokaryotic RNA Polymerase
Structure and Components
Prokaryotic RNA polymerase is typically composed of five core subunits, forming a complex known as the holoenzyme. This includes two alpha (α) subunits, one beta (β) subunit, one beta-prime (β’) subunit, and a sigma (σ) factor that is essential for the initiation of transcription.
Mechanism of Action
The action of prokaryotic RNA polymerase begins with the binding of the sigma factor to the promoter region of DNA. This step ensures the enzyme’s correct positioning for the start of RNA synthesis. The polymerase then unwinds a short segment of the DNA to read the template strand and begin synthesizing RNA, a process that continues until a termination signal is reached.
Eukaryotic RNA Polymerase
Structure and Components
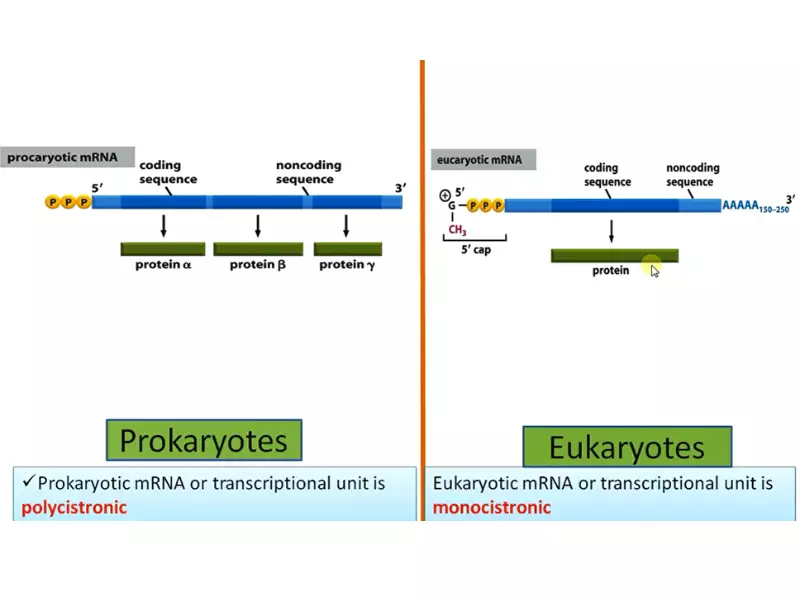
Eukaryotic cells contain three distinct RNA polymerases (I, II, and III), each responsible for transcribing different types of genes. RNA polymerase II, for example, is primarily involved in the synthesis of messenger RNA (mRNA). These enzymes are much larger and more complex than their prokaryotic counterparts, consisting of up to 12 or more subunits.
Mechanism of Action
The mechanism of action for eukaryotic RNA polymerase also involves the binding to promoter regions but requires a host of additional proteins called transcription factors to initiate the process. Transcription by eukaryotic polymerases includes not only the elongation of the RNA chain but also extensive processing of the RNA molecule, such as capping, splicing, and polyadenylation.
Comparative Analysis
Structural Variations
Core Enzyme Structure
The core enzyme structure of prokaryotic RNA polymerase, while simpler, is functionally similar to that of eukaryotic polymerases in its ability to synthesize RNA. However, the eukaryotic enzymes’ complexity, with multiple forms and numerous subunits, reflects the higher level of regulatory control required for gene expression in these organisms.
Regulatory Subunits
The presence of the sigma factor in prokaryotes and the requirement for multiple transcription factors in eukaryotes highlight the differences in regulatory mechanisms governing transcription initiation. These components ensure that RNA polymerase accurately and efficiently initiates transcription at the correct gene locations.
Functional Differences
Initiation Process
The initiation of transcription involves distinct processes in prokaryotic and eukaryotic cells. In prokaryotes, the sigma factor plays a crucial role in recognizing promoter regions. In contrast, eukaryotic transcription initiation requires a complex assembly of transcription factors and coactivators to recruit the polymerase to the promoter.
Elongation and Termination
During elongation, RNA polymerase moves along the DNA template, synthesizing RNA. This phase is relatively similar in both cell types, although specific regulatory proteins can modulate the process. Termination in prokaryotes often involves rho-dependent or rho-independent signals, whereas eukaryotic termination involves cleavage of the newly synthesized RNA and additional processing steps.
Regulatory Mechanisms
Promoter Recognition
Promoter recognition is a key step in transcription initiation. Prokaryotic RNA polymerase utilizes the sigma factor for this purpose, while eukaryotic cells deploy a complex array of transcription factors and mediator proteins to ensure precise binding of RNA polymerase to the DNA.
Response to Cellular Signals
Both prokaryotic and eukaryotic cells have evolved mechanisms to regulate transcription in response to internal and external signals. In prokaryotes, this can involve the activation or repression of sigma factors. In eukaryotes, the modulation of transcription factor activity and post-translational modifications of RNA polymerase subunits play significant roles in responding to cellular cues.
Biological Implications
Gene Expression
Impact on Transcription
The differences between prokaryotic and eukaryotic RNA polymerase have profound implications for gene expression. In prokaryotes, the simpler structure and the direct involvement of sigma factors allow for a swift response to environmental changes, which is crucial for survival in varying conditions. Eukaryotic cells, with their complex regulatory mechanisms involving multiple RNA polymerases and transcription factors, can finely tune gene expression in response to developmental cues and external signals. This regulation is essential for the proper functioning of multicellular organisms, ensuring that genes are expressed at the right time, place, and amount.
Adaptation and Evolution
Evolutionary Significance
The evolution of RNA polymerase from prokaryotes to eukaryotes marks a significant point in the history of life. The increased complexity of eukaryotic RNA polymerase and its regulatory mechanisms reflects the evolutionary pressure to support more complex cellular functions and multicellular life. This complexity allows for the diverse forms of life we see today, each adapted to their unique environment through the intricate control of gene expression.
Research and Biotechnology
Applications in Genetic Engineering
The understanding of RNA polymerase function in both prokaryotic and eukaryotic organisms has paved the way for groundbreaking advancements in genetic engineering. Techniques such as CRISPR-Cas9 gene editing rely on precise control over gene expression, which is only possible with a deep understanding of transcription mechanisms. The ability to modify gene expression has vast applications, from developing drought-resistant crops to treating genetic disorders in humans.
Challenges and Future Directions
Research Challenges
Current Limitations in Study
Despite the advancements in our understanding of RNA polymerase and its role in gene expression, several challenges remain. One major obstacle is the complexity of eukaryotic transcription, involving an array of coactivators, mediators, and transcription factors. Additionally, the dynamic nature of transcription, with its rapid initiation and termination phases, makes it difficult to capture and study these processes in real-time. Moreover, the interaction between RNA polymerase and DNA is influenced by the chromatin structure, which adds another layer of complexity to understanding transcription regulation in eukaryotes.
Future Research
Potential Areas of Exploration
Future research in the field of RNA polymerase and gene expression is poised to tackle these challenges with innovative approaches. Key areas include:
- Single-molecule imaging techniques to visualize transcription in real-time, providing insights into the dynamics of RNA polymerase action.
- Advancements in computational biology to model the complex interactions between RNA polymerase, DNA, and regulatory proteins, offering predictions that can guide experimental research.
- Epigenetics research to understand how modifications of the chromatin landscape influence RNA polymerase access and gene expression.
- Synthetic biology approaches to redesign RNA polymerase and its associated factors for specific applications in medicine and biotechnology, such as developing new therapies for controlling gene expression in diseases.
Frequently Asked Questions
What is RNA Polymerase?
RNA polymerase is an essential enzyme in the process of transcription, where it reads DNA templates to synthesize messenger RNA (mRNA). This mRNA then serves as a template for protein synthesis in the cell, making RNA polymerase critical for gene expression and the regulation of cellular functions.
How do Prokaryotic and Eukaryotic RNA Polymerases Differ?
The main difference between prokaryotic and eukaryotic RNA polymerases is in their structure and the complexity of their regulatory mechanisms. Prokaryotes have a single RNA polymerase that handles all RNA synthesis, while eukaryotes have three distinct types of RNA polymerases (I, II, and III), each specialized for transcribing different types of genes. Additionally, eukaryotic RNA polymerases require various transcription factors to initiate transcription, unlike their prokaryotic counterparts.
Why are Eukaryotic RNA Polymerases More Complex?
Eukaryotic RNA polymerases are more complex due to the compartmentalization of eukaryotic cells and the sophisticated control needed over gene expression. This complexity allows for the precise regulation of gene expression in response to various internal and external signals, a necessity for the development and maintenance of multicellular organisms.
What Role do Transcription Factors Play in Eukaryotic Transcription?
Transcription factors are proteins that help eukaryotic RNA polymerases recognize and bind to specific DNA sequences called promoters, which are located at the beginning of genes. These factors are crucial for initiating transcription in eukaryotic cells, as they mediate the interaction between RNA polymerase and the DNA template, ensuring that genes are expressed at the right time and in the right amount.
Conclusion
The distinction between prokaryotic and eukaryotic RNA polymerase is a testament to the evolutionary complexity of life. This divergence not only reflects the differing requirements of gene expression and regulation in simple versus complex organisms but also highlights the sophistication inherent in eukaryotic cells. Understanding these differences deepens our appreciation of the molecular mechanisms that govern life and lays the groundwork for advancements in biotechnology and medicine.
In exploring the nuances between prokaryotic and eukaryotic RNA polymerases, we gain insights into the broader themes of biology, including the evolution of cellular life and the intricate dance of gene expression. Such knowledge is crucial for the development of new therapies and technologies, underscoring the importance of basic science in paving the way for future innovations.
