Genome sequencing, a process that uncovers the complete DNA sequence of an organism’s genome, has revolutionized the fields of biology and medicine. This sophisticated procedure provides insights into genetic disorders, evolutionary biology, and the mechanisms of diseases. As technology has advanced, two primary methods have emerged as frontrunners in the quest to decode genetic information: hierarchical sequencing and whole genome shotgun sequencing.
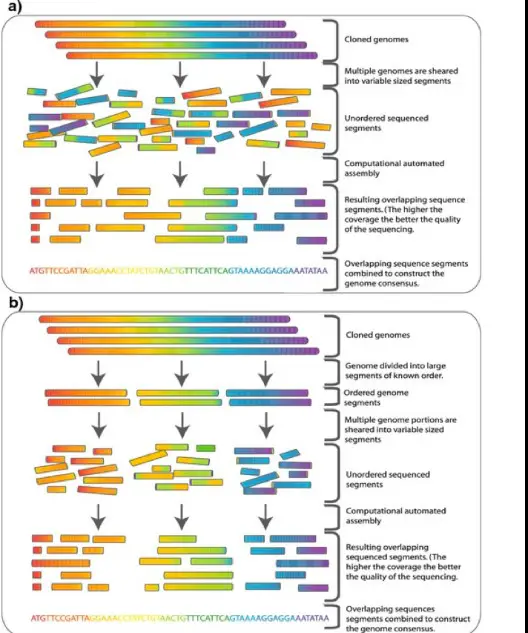
The difference between hierarchical and whole genome shotgun sequencing lies in their approach to unraveling the genome’s complex structure. Hierarchical sequencing, also known as clone-by-clone sequencing, involves mapping and selectively sequencing specific regions of the genome. In contrast, whole genome shotgun sequencing breaks the entire genome into small pieces, sequences these fragments in a random order, and then uses computational strategies to assemble the pieces back together.
While hierarchical sequencing offers high accuracy and is particularly suited for large genomes, it is time-consuming and resource-intensive. On the other hand, whole genome shotgun sequencing is faster and more cost-effective but comes with its own set of challenges, including complexity in assembly and potential for errors. Each method has contributed significantly to our understanding of genetic blueprints, influencing both scientific research and the development of medical treatments.
Genome Sequencing Basics
What is Genome Sequencing?
Definition and Purpose
Genome sequencing is the process of determining the exact sequence of nucleotides in an organism’s DNA. This sequence includes all four bases: adenine (A), guanine (G), cytosine (C), and thymine (T). The purpose of genome sequencing is to unlock the genetic blueprint of an organism. This information is crucial for understanding genetic variations, diagnosing genetic disorders, advancing personalized medicine, and enhancing our comprehension of evolutionary biology.
Key Technologies
Brief History
The journey of genome sequencing began in the 1970s with the development of the Sanger sequencing method. This method, also known as the chain termination method, was a groundbreaking advancement that paved the way for the first sequencing of the human genome. Over the decades, sequencing technologies have evolved rapidly, transitioning from labor-intensive methods to high-throughput technologies that can sequence entire genomes quickly and cost-effectively.
Current Technologies
Today, two main technologies dominate the field of genome sequencing: Next-Generation Sequencing (NGS) and Third-Generation Sequencing. NGS technologies, such as Illumina sequencing, can sequence billions of DNA strands simultaneously, dramatically reducing the time and cost of sequencing. Third-Generation Sequencing, including platforms like PacBio and Oxford Nanopore, offers even longer read lengths, allowing for more detailed analysis of genomic regions.
Hierarchical Sequencing
Overview
Definition and Process
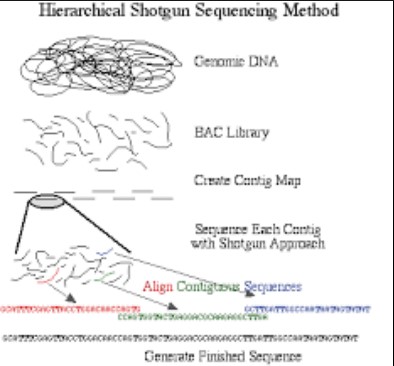
Hierarchical sequencing, or clone-by-clone sequencing, is a methodical approach that organizes the genome into manageable sections before sequencing. This process involves creating a physical map of the genome, then systematically sequencing selected portions. This approach is known for its precision and reliability in decoding complex genomes.
Key Steps
- Mapping: The first step involves creating a physical map of the genome, identifying and categorizing different regions.
- Cloning: Selected regions of the genome are then cloned into vectors to create copies for sequencing.
- Sequencing: Each cloned segment is sequenced individually, using methods such as Sanger sequencing.
- Assembly: Finally, the sequenced fragments are meticulously assembled back into the complete genome sequence.
Advantages
- Accuracy: Hierarchical sequencing is renowned for its high accuracy, minimizing errors in the final genome sequence.
- Suitability for Large Genomes: Its organized approach makes it especially suitable for sequencing large, complex genomes, such as those of humans and plants.
Challenges
- Time-consuming: This method requires significant time due to its step-by-step approach.
- Resource-intensive: The need for physical mapping and cloning makes hierarchical sequencing a resource-heavy process.
Whole Genome Shotgun Sequencing
Overview
Definition and Process
Whole Genome Shotgun (WGS) sequencing involves randomly breaking down the entire genome into smaller fragments, sequencing these fragments without prior mapping, and then using computational tools to reassemble the sequence. This method bypasses the need for physical mapping, offering a faster route to sequencing entire genomes.
Key Steps
- Fragmentation: The genome is randomly broken into small pieces.
- Sequencing: Each fragment is sequenced using high-throughput technologies.
- Assembly: Computational algorithms are used to align and merge the sequences into the full genome sequence.
Advantages
- Speed: WGS sequencing significantly reduces the time required to sequence entire genomes.
- Cost-effectiveness: By eliminating the need for mapping and cloning, WGS is more cost-effective compared to hierarchical sequencing.
Challenges
- Complexity in Assembly: The lack of a physical map can make the assembly process complex, especially with genomes that have repetitive sequences.
- Errors and Gaps: The possibility of errors and gaps in the assembled genome is higher with WGS, requiring sophisticated computational tools to identify and correct these issues.
Comparative Analysis
Accuracy and Reliability
When comparing hierarchical sequencing and whole genome shotgun sequencing (WGS), it’s crucial to consider accuracy and reliability. Hierarchical sequencing, with its methodical approach, tends to have lower error rates. This precision is due to the meticulous mapping and cloning steps that ensure each part of the genome is correctly sequenced and assembled. In contrast, WGS, despite its advancements, can face challenges with repetitive sequences that may lead to errors or gaps in the assembled genome. Thus, for projects where accuracy is paramount, hierarchical sequencing often holds the edge.
Time and Cost
The time and cost involved in genome sequencing are significantly influenced by the chosen method. Hierarchical sequencing, while highly accurate, is time-consuming and costly due to its elaborate preparatory steps. WGS, on the other hand, stands out for its speed and cost-effectiveness, leveraging high-throughput sequencing technologies that bypass the need for extensive mapping and cloning. This makes WGS a more appealing option for projects with limited budgets or tight timelines.
Suitability
Deciding which method to use depends largely on the project’s specific needs:
- Large, complex genomes: Hierarchical sequencing is often the better choice due to its detailed mapping, which can navigate complex genetic landscapes.
- Rapid, cost-effective sequencing: WGS is ideal for projects requiring quick turnaround times or for organisms with smaller, less complex genomes.
Technological Advances
Technological advances in sequencing have significantly impacted the choice between hierarchical and WGS methods. The advent of Next-Generation Sequencing (NGS) technologies has made WGS more feasible and accurate, narrowing the accuracy gap between the two methods. As sequencing technologies continue to evolve, the decision-making process will increasingly consider factors such as read length, throughput, and the ability to handle repetitive sequences.
Case Studies
Human Genome Project
The Human Genome Project (HGP), one of the most ambitious biological research projects in history, primarily utilized hierarchical sequencing. This approach allowed scientists to meticulously map and sequence the human genome with remarkable accuracy, laying the foundation for future genetic research and medical breakthroughs. The project’s success highlighted the value of hierarchical sequencing in handling large, complex genomes.
Microbial Genomics
In the realm of microbial genomics, WGS has demonstrated significant success. The smaller size and less complex nature of microbial genomes make them ideal candidates for WGS, allowing for rapid sequencing and analysis. This method has enabled the identification and understanding of numerous microbial species, contributing to advances in medicine, environmental science, and biotechnology.
Future Directions
Emerging Technologies
The future of genome sequencing looks toward emerging technologies that promise even greater speed, accuracy, and affordability. Third-Generation Sequencing technologies, such as single-molecule sequencing, offer longer read lengths and the ability to sequence without amplification, reducing the potential for errors. These advancements are expected to further blur the lines between hierarchical and WGS, potentially leading to new sequencing paradigms.
Challenges and Opportunities
The advancements in sequencing technologies bring both challenges and opportunities, particularly in data analysis and storage. The sheer volume of data generated by modern sequencing methods requires robust computational tools for assembly, analysis, and storage. This necessitates ongoing development in bioinformatics to keep pace with the evolving landscape of genome sequencing.
The transition towards more advanced sequencing technologies presents an opportunity to tackle previously intractable genomic regions and to apply genomic insights across a broader range of disciplines. However, the increasing complexity and volume of sequencing data also demand enhanced computational resources and innovative data management solutions. As the field progresses, the balance between technological capabilities and the practicalities of data handling will play a pivotal role in shaping the future of genomics research.
Frequently Asked Questions
What is Genome Sequencing?
Genome sequencing is the process of determining the exact sequence of nucleotides (A, T, C, G) in an organism’s DNA. This comprehensive mapping allows scientists to understand genetic variations, identify mutations, and explore genomic structures that contribute to specific traits or diseases.
Why is Genome Sequencing Important?
Genome sequencing has critical applications in various fields, including medicine, agriculture, and environmental science. It enables the identification of genetic disorders, the development of targeted therapies, and the improvement of crop varieties. Furthermore, it provides invaluable insights into evolutionary biology and the functioning of ecosystems.
How Does Hierarchical Sequencing Differ from Whole Genome Shotgun Sequencing?
Hierarchical sequencing is a methodical approach that involves mapping the genome and sequencing designated sections sequentially. It is highly accurate but time-consuming. Whole genome shotgun sequencing, conversely, fragments the genome into many pieces, sequences these fragments simultaneously, and then assembles the sequences into a complete genome, offering speed and efficiency at the potential cost of accuracy.
What Are the Challenges of Whole Genome Shotgun Sequencing?
The main challenges of whole genome shotgun sequencing include the complexity of assembling the sequenced fragments back into a complete genome. This process can be complicated by repetitive sequences and can result in gaps or errors in the assembled genome, requiring sophisticated computational tools to resolve.
Conclusion
Choosing between hierarchical and whole genome shotgun sequencing involves balancing accuracy, time, and cost, each method catering to different research needs and resources. As genomic sequencing technologies continue to evolve, the decision will increasingly depend on the specific requirements of the project at hand.
The future of genome sequencing looks promising, with advancements in technology paving the way for faster, more accurate, and more affordable methods. These developments will undoubtedly expand our understanding of genetics, further revolutionizing medicine, agriculture, and biology. As we continue to unravel the mysteries of the genome, the choice between hierarchical and whole genome shotgun sequencing will remain a critical consideration in the quest to decode life’s blueprint.